Revista Brasileira de Ginecologia e Obstetrícia. 2015;37(2):94-99
The aim of this study was to compare the performance of two human papillomavirus (HPV) genotyping techniques, Linear Array and PapilloCheck, in women with high-grade squamous intraepithelial lesion (HSIL).
A total of 88 women with cytological diagnosis of HSIL were recruited at 2 reference centers in cervical pathology in Salvador, Bahia, Brazil, from July 2006 to January 2009. After the cytological diagnosis of HSIL, cervix cells were collected to determine the HPV genotype and a biopsy was obtained under colposcopic vision for histopathological analysis. After the confirmation of CIN2+ by histopathology, HPV genotyping was performed on 41 women by the Linear Array and PapilloCheck methods.
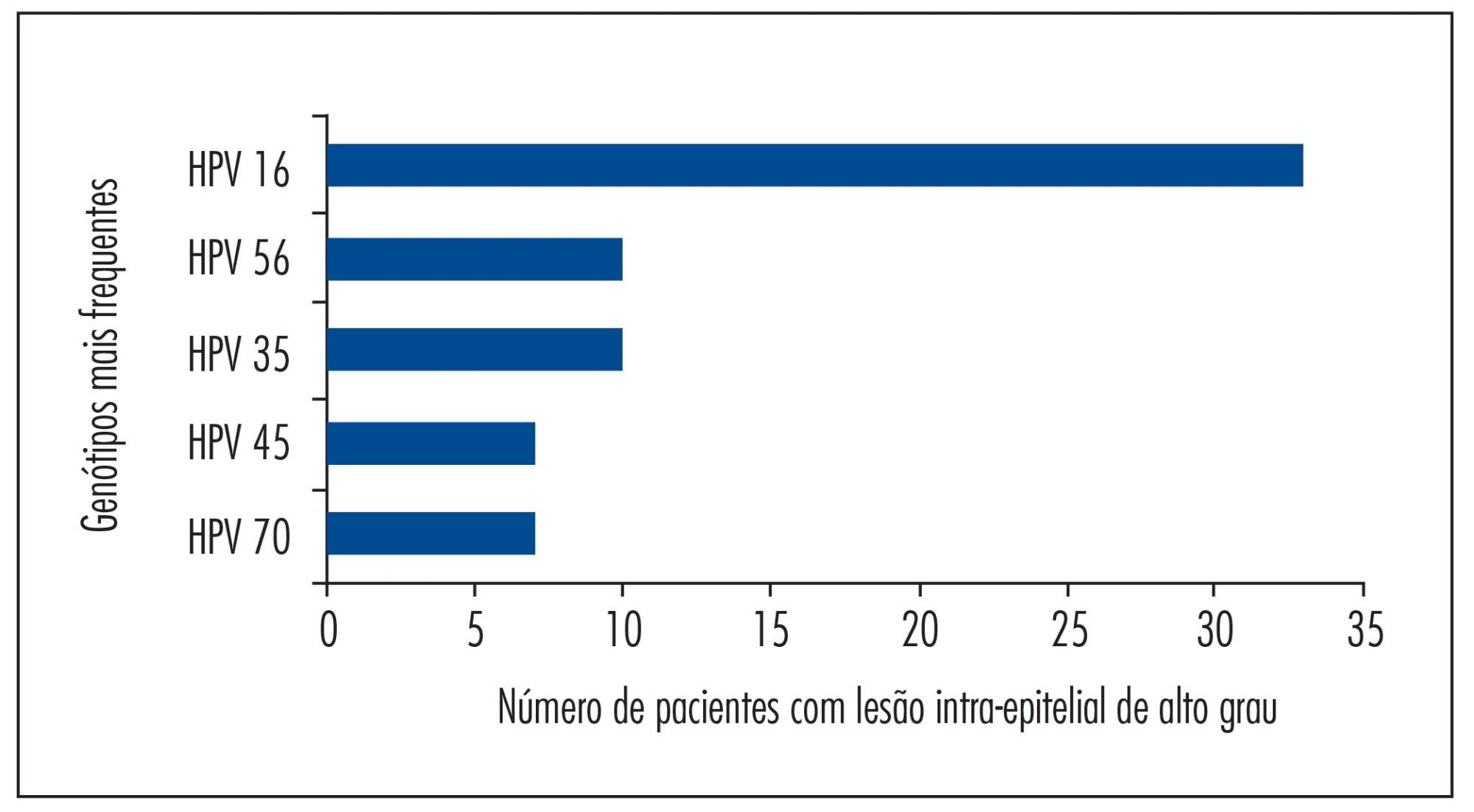
Both tests showed an overall concordance rate for HPV detection of 97.2% (35/36). Of the 36 valid samples, 35 (97.2%) were positive in both tests and 1 (2.8%) was discordant, with the Linear Array indicating the presence of multiple types. The most prevalent HPV genotypes detected by the Linear Array technique were HPV 16, HPV 56, HPV 35, HPV 45, and HPV 70; and those detected by the PapilloCheck technique were HPV 16, HPV 56, HPV 11, HPV 35, and HPV 42. A similar rate of infection with multiple HPV types was observed with the two tests (72.5% with the Linear Array and 75.0% with the PapilloCheck).
Linear Array genotyping assay and PapilloCheck showed equivalent performance for the detection of oncogenic HPV types in women with HSIL, with PapilloCheck having the advantage of being a method that avoids subjectivity when reading the HPV genotypes.
Search
Search in:


Comments